1. Data Integration and Standardization
Integrating datasets from EHRs, genetic data, lab results, and patient-reported outcomes was challenging due to varying formats and qualities. Mismatched medical codes like ICD-10 and SNOMED CT caused inconsistencies, while missing data and conflicting details required resolution and tracking. These issues complicated the creation of a clean, unified dataset for analysis.
2. Patient Variability and Subgroup Representation
Differences in demographics (age, gender, ethnicity), medical histories (comorbidities, disease severity), medications, and lifestyle factors (e.g., smoking, activity levels) made accounting for variability challenging. Hidden factors, such as undiagnosed conditions or environmental influences, introduced biases that could distort results.
3. Patient Grouping and Similarity Analysis
Creating meaningful patient groupings was difficult due to variations in medical histories, lab results, genetic markers, symptoms, and social factors like income and access to care. Many patients didn’t fit neatly into a single group, making clustering methods like k-means and hierarchical clustering challenging to implement effectively.
4. Reliable and Reproducible Results
Ensuring reliable results required handling complex data and addressing uncertainties. Missing data and confounding variables posed significant challenges, demanding advanced techniques like survival analysis and mixed-effects models. Probabilistic predictions added complexity, requiring external validation to confirm relevance in real-world scenarios.
5. Treatment Risk Analysis and Impact Assessment
Evaluating treatment risks was challenging due to differences in patient demographics, pre-existing conditions, concurrent medications, and external factors like seasonal trends and socioeconomic disparities. Issues like inconsistent adherence and variable data collection further complicated efforts to ensure findings reflected real-world conditions.
To address the challenges of evaluating hydroxychloroquine safety, we developed a comprehensive solution that integrates advanced data processing, analysis, and modeling techniques. The product offers tools to:
1. Integrate and Standardize Diverse Medical Data:
Consolidates large datasets from multiple sources (EHRs, genetic data, lab results, patient-reported outcomes) into the OMOP Common Data Model (CDM), ensuring consistency and compatibility.
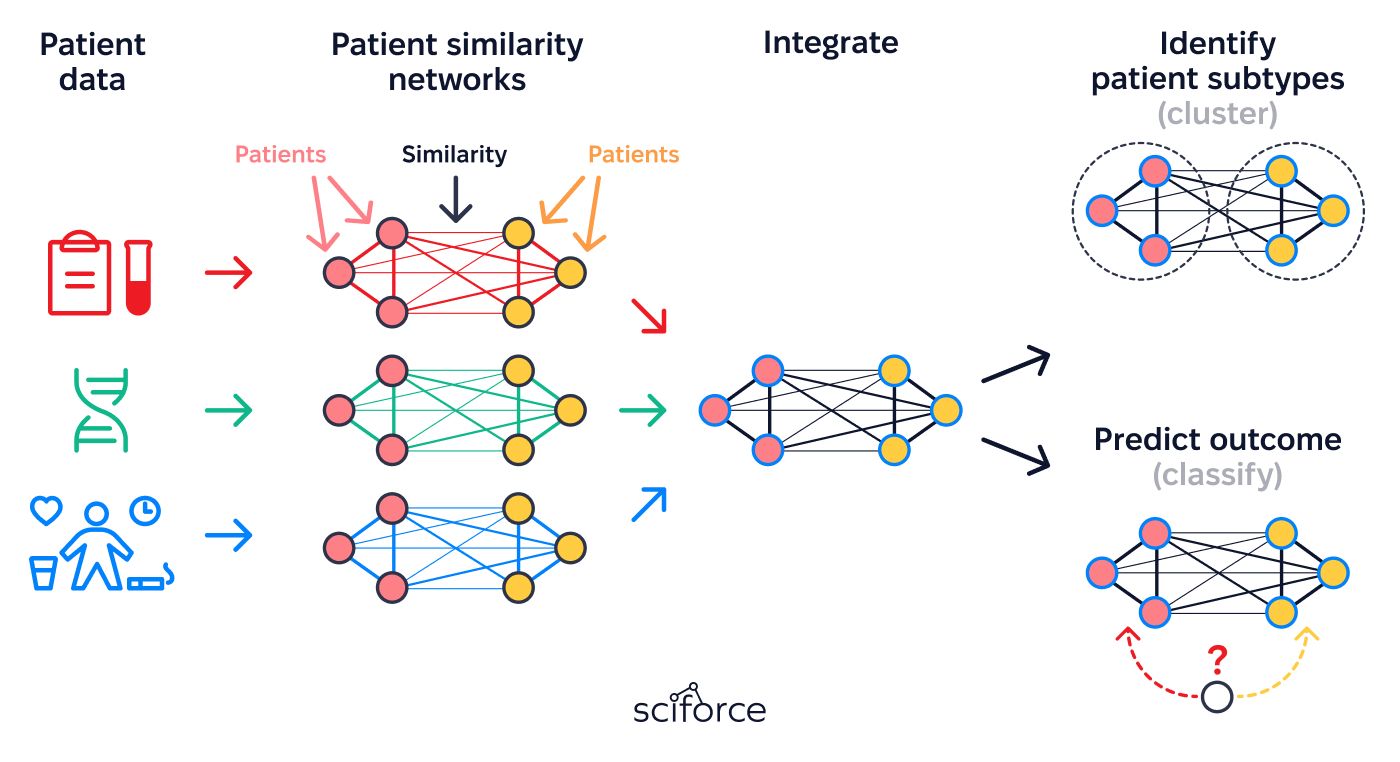
2. Build Patient Similarity Networks (PSNs):
Groups patients with similar characteristics (clinical, genetic, and phenotypic) to enhance risk analysis and treatment outcome prediction.
3. Analyze Risks and Long-Term Safety:
Provides detailed insights into treatment risks and safety, accounting for variability across patient subgroups and external factors.
4. Support Research and Decision-Making:
Enables evidence-based findings for scientific publications and regulatory reports, facilitating informed clinical decisions.
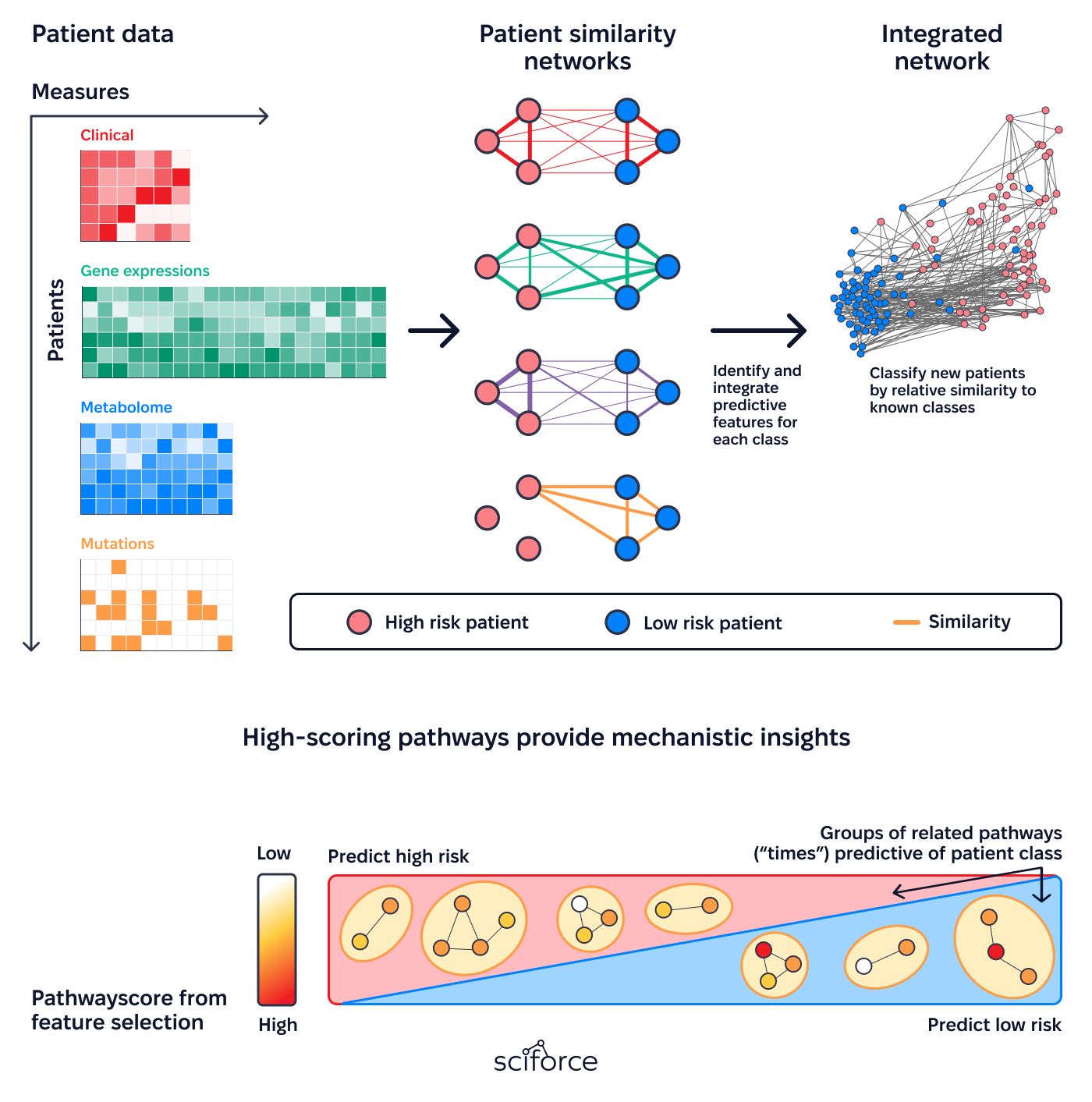
1. Unified Patient Network Framework
Creates a streamlined system for identifying patient similarities by combining clinical histories, genetic data, phenotypic traits, and social determinants of health (SDOH). It integrates this information from various sources into a unified network that supports precise and meaningful analysis.
2. Context-Aware Clustering Algorithms
Uses advanced clustering methods like hierarchical, density-based, and k-means to group patients accurately. These algorithms combine data such as clinical histories, genetic markers, phenotypic traits, and SDOH to create detailed and practical health profiles.
3. Confounder-Resilient Matching System
Uses methods like propensity score matching, inverse probability weighting, and stratification to carefully account for confounders. These techniques reduce bias, improve fairness in comparisons, and ensure results are precise and relevant for clinical decisions.
4. Dynamic Sensitivity Exploration
Performs thorough sensitivity analyses, including E-values, tipping point scenarios, and leave-one-out tests, to ensure findings are stable and reliable. It highlights potential weaknesses caused by unmeasured confounders, checks the validity of model assumptions, and adjusts methods to address variations and complexities in real-world data.
5. Insight-Enriched Clinical Tools
Features interactive dashboards for subgroup-specific risk assessments, predictive models for treatment outcomes, scenario-driven decision support tools, detailed effectiveness reports, and a proactive alert system for potential adverse events.
Iterative Methodology Refinement
Adaptation to Rheumatoid Arthritis Patients
Multidisciplinary Team Coordination
Addressing Key Data Challenges
Data Integration and Standardization:
OMOP CDM:
Standardizes and integrates diverse medical data, including EHRs, laboratory results, and genetic profiles, ensuring consistency and compatibility for robust analysis.
OHDSI Tools:
Data Processing, Analysis, and Sensitivity Assessment:
Python Libraries:
R Packages:
SQL:
Visualization and Dashboard Development:
Python Libraries:
Deployment and Hosting:
AWS:
Starting Point (Point A):
At the beginning of the project, the client faced significant challenges:
Final Outcome (Point B):
The project delivered an integrated solution centered on Patient Similarity Networks (PSNs), providing: